Designing Proteins to be Molecular Machines
Table of contents

Why our society chooses to idolize movie stars would be a complete mystery to visiting aliens. What’s so admirable about spending your life pretending to be someone else? In most cases, the people who actors and actresses attempt to portray aren’t even real people to begin with. Mankind would be in a much better place if Nobel Prize winners were household names instead. Then, we wouldn’t have to introduce Richard Smalley, the man who won a Nobel Prize for the discovery of fullerenes.
In the early 2000s, Mr. Smalley engaged in an open debate with Eric Drexler – author of Engines of Creation and There’s Plenty of Room At The Bottom – about the theoretic feasibility of self-assembly. Back then, many people envisioned nanobots as tiny little nano-machines that could assemble more of themselves, and gray goo theory concerned itself with what might happen if the machines started to replicate uncontrollably. Twenty years later, and we’re now creating molecular machines never before seen in nature.
All organisms on Earth employ the same workforce to perform a wide range of essential biochemical tasks. This workforce is comprised of proteins, which are constructed from a long string of amino acids attached to each other.
Credit: Space.com
The Most Amazing Machines in the World
Someone else who ought to be a household name is serial entrepreneur David Baker, a biochemist who recently gave a TED talk on the most amazing machines in the world – proteins – which are responsible for almost everything that happens in biology.
In that talk, Mr. Baker discussed how the shapes of proteins help them accomplish their remarkable functions which are completely specified by the sequence of 20 amino acid building blocks used to create proteins. (Nobody knows why nature chose these 20 specific amino acids out of thousands of possible selections.) Chemical forces between these stringy molecules of chained amino acids cause them to fold up into 3D shapes in a fraction of a second, forming proteins.
Tinker with the amino acids and you can manipulate the proteins, but it’s easier said than done. Just to get an idea of the complexity involved here, protein modeling algorithms need to consider more combinatorial sequences than there are atoms in the universe. That’s why studying proteins has been historically difficult, and we’ve only been able to manipulate existing proteins in small ways.
Today, all that’s changed thanks to technologies like deep learning which help us decipher how proteins work, and gene editing which allows us to create synthetic genes which in turn create entirely artificial proteins with desirable properties. It hints at a future where designer proteins will be the norm.
“In 10 to 15 years, I believe that nearly all the proteins used in technology will be designed de novo to have the properties that are wanted, rather than properties adapted from something that happened accidentally during evolution.”
Credit: David Baker, IPD
The term de novo is simply another way of saying “built from the ground up,” and it all happens using computational protein design software that creates proteins in the digital world before the real world. The total number of proteins found in nature is minuscule compared to the total number of proteins that can be made using any combination of amino acids. Below you can see a yellow grid with a little grey square on it.
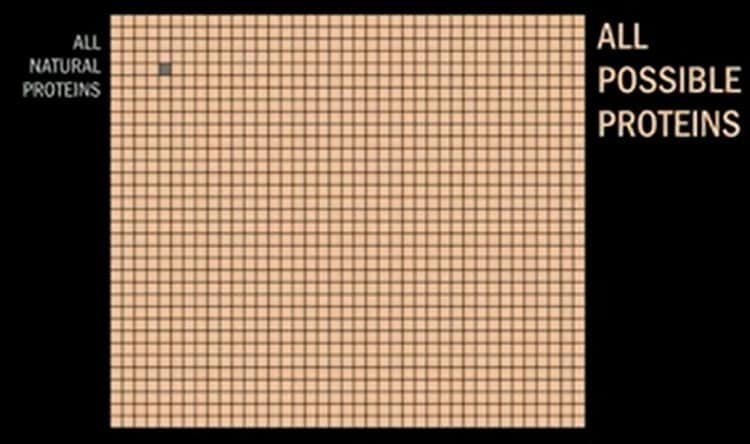
The grey square seen above represents all proteins found in nature. The rest of the little squares represent all possible proteins. The broad number of applications to be addressed is something that’s attracting a lot of attention from big pharma.
Creating Novel Molecular Machines

A few years ago we looked at 6 Protein Engineering Startups working on designing proteins to do cool stuff. One of those was Synthorx, a company that added the DNA letters X and Y so that cells can make proteins with up to 172 different amino acids versus the 20 amino acids created by Mother Nature. It sounds incredible because it is, and that’s why Sanofi outbid a number of other suitors to acquire Synthorx for $2.5 billion late last year. What Sanofi acquired were “optimized biologics” called Synthorins which are expected to address the shortcomings of some existing cancer treatments.
Synthorx isn’t the only company creating molecular machines never before seen in nature.
The Institute of Protein Design
Founded in 2012, the University of Washington Institute for Protein Design (IPD) is funded by at least $56 million in grants with the money being spent on designing “a new world of proteins to address 21st-century challenges in medicine, energy and technology.” In 2017, researchers at IPD managed to design four new synthetic protein folds never before observed in nature. Today, they’re working with industry leaders like Amgen to create new proteins with novel properties using the process described below:

IPD is considered to be the world leader in creating new proteins with new functions from scratch, and multiple startups have been spun out of their research. The main man behind these spinouts is the same gentleman who gave the aforementioned TED talk on protein engineering, David Baker, who also happens to be Director of the IPD. His success stories include PvP Biologics, a startup that took in $35 million in funding before being acquired this spring by Takeda for $330 million. PvP was working on a treatment for celeriac disease, which means all you gluten-free eaters may have to find something else to become unnecessarily worried about.
Let’s look at a few more companies that have been spun out of IPD.
The First De Novo Protein Therapeutic

Spun out in 2019, Neoleukin Therapeutics (NLTX) is developing proteins that demonstrate specific pharmaceutical properties that provide potentially superior therapeutic benefit over native proteins. They’re now a $440 million publicly traded company that’s working on the world’s first computationally-designed de novo protein therapeutic. Called NL-201, it’s positioned as a potential best-in-class IL-2/IL-15 cancer immunotherapy. Drug development is a particularly risky exercise, so expect the share price to exhibit lots of volatility as additional information becomes available as to their progress.
Software Eats Proteins
Also spun out of IPD is Seattle startup Cyrus Biotechnology which has taken in $11.3 million in disclosed funding to commercialize the world’s leading protein engineering platform – Rosetta – which is the first software experimentally proven to design new proteins computationally. First built in the late 90s, Rosetta was being used internally by IPD when they decided to commercialize it for use externally by companies and academics. The end result is Cyrus Bench, an easy-to-use version of the Rosetta molecular modeling and protein design software package.
While half of their customers have never even purchased modeling software before, the other half includes names like BASF, Harvard Medical School, Janssen Pharmaceuticals, and Illumina.
10,000 Proteins a Week
Another IPD spin out, Arzeda, has taken in $15.2 million in funding so far. They call themselves “the protein design company” which is a fitting name considering they design, build, and test 10,000 proteins a week for applications ranging from agtech microbiomes to food sweeteners. In particular, they’re working with enzymes, proteins that act as biological catalysts which are used to accelerate chemical reactions. Sounds quite familiar to the work going on over at Zymergen and Ginkgo Bioworks, two of the most exciting synthetic biology startups there are today.
Another startup that’s examining the behavior of proteins on a large scale is A-Alpha Bio, also a spinout from IPD.
Solving the Library to Library Problem
Founded in 2017, Seattle startup A-Alpha Bio started out with grants from the National Science Foundation which led to a proper seed round in Sept 2019 led by moonshot investor OS Fund. The startup has now taken in $4.4 million in disclosed funding to develop AlphaSeq, a synthetic biology platform used to accelerate drug development by enabling library-on-library measurements of protein interactions. The problem involves combing through millions of protein interactions to find the ones which lead to viable therapeutic discoveries. An article by GeekWire likens the “library to library” problem to having millions of locks in one pile, and millions of keys in another pile, then trying to figure out which keys open which locks.
Just because a technology like protein design has great potential doesn’t mean every startup will succeed. One protein engineering startup we looked at before was IPD spin out Virvio. Since our article in April 2018, Virvio’s website has disappeared leading us to believe they’ve gone pear-shaped. Prior to their disappearance, Virvio was looking to design proteins to combat influenza.
Lastly, for all of you armchair Twitter CEOs out there, you too can help to solve some of mankind’s biggest problems and have fun doing so. Baker Lab at the University of Washington has developed a protein-folding game you can play which helps scientists better understand proteins, something far more noble – and presumably more fun – than boycotting brands for perceived indiscretions.
Conclusion
A few themes stand out among these startups, the first being the use of artificial intelligence to advance the development and understanding of proteins. The second is that all these startups seem to start out with an unlimited number of use cases across all industries, but eventually end up gravitating towards pharmaceutical applications. It makes sense considering how artificial intelligence has seen the most initial use cases in healthcare. Rest assured, this is only the tip of the iceberg as some of the world’s most elite scientists work to evolve and improve upon the most amazing molecular machines in the world.
Sign up to our newsletter to get more of our great research delivered straight to your inbox!
Nanalyze Weekly includes useful insights written by our team of underpaid MBAs, research on new disruptive technology stocks flying under the radar, and summaries of our recent research. Always 100% free.